This vignette demonstrates how a GAM model changes with increased flexibility.
The data we will use here contains tract profiles from diffusion MRI measurements in a group of patients with Amyotrophic Lateral Sclerosis (ALS) and a group of matched controls (Sarica et al. 2017).
We start by loading the tractable library and
gratia, as well as ggplot2, which we will use
to interpret the fitted model.
Next, we will use a function that is included in
tractable to read this dataset directly into memory.
Importantly, both the group (“ALS” or “CTRL”) and the subject identifier (“subjectID”) need to be factors for subsequent analysis to work properly.
df_sarica <- read_afq_sarica(na_omit = TRUE)
df_sarica
#> # A tibble: 93,377 × 8
#> subjectID tractID nodeID fa md age group gender
#> <fct> <chr> <dbl> <dbl> <dbl> <dbl> <fct> <fct>
#> 1 subject_000 Left Thalamic Radiation 0 0.174 0.965 54 ALS F
#> 2 subject_000 Left Thalamic Radiation 1 0.227 0.912 54 ALS F
#> 3 subject_000 Left Thalamic Radiation 2 0.282 0.874 54 ALS F
#> 4 subject_000 Left Thalamic Radiation 3 0.318 0.851 54 ALS F
#> 5 subject_000 Left Thalamic Radiation 4 0.341 0.830 54 ALS F
#> 6 subject_000 Left Thalamic Radiation 5 0.353 0.812 54 ALS F
#> 7 subject_000 Left Thalamic Radiation 6 0.357 0.802 54 ALS F
#> 8 subject_000 Left Thalamic Radiation 7 0.357 0.795 54 ALS F
#> 9 subject_000 Left Thalamic Radiation 8 0.359 0.790 54 ALS F
#> 10 subject_000 Left Thalamic Radiation 9 0.371 0.784 54 ALS F
#> # ℹ 93,367 more rowsWe fit models with different levels of flexibility, encoded in
different values of k:
k_values <- c(4, 8, 16, 32)
models <- list()
for (i in 1:length(k_values)){
models[[i]] <- tractable_single_tract(
target = "fa",
df = df_sarica,
tract = "Right Corticospinal",
regressors = c("age", "group"),
node_k = k_values[i],
node_group = "group"
)
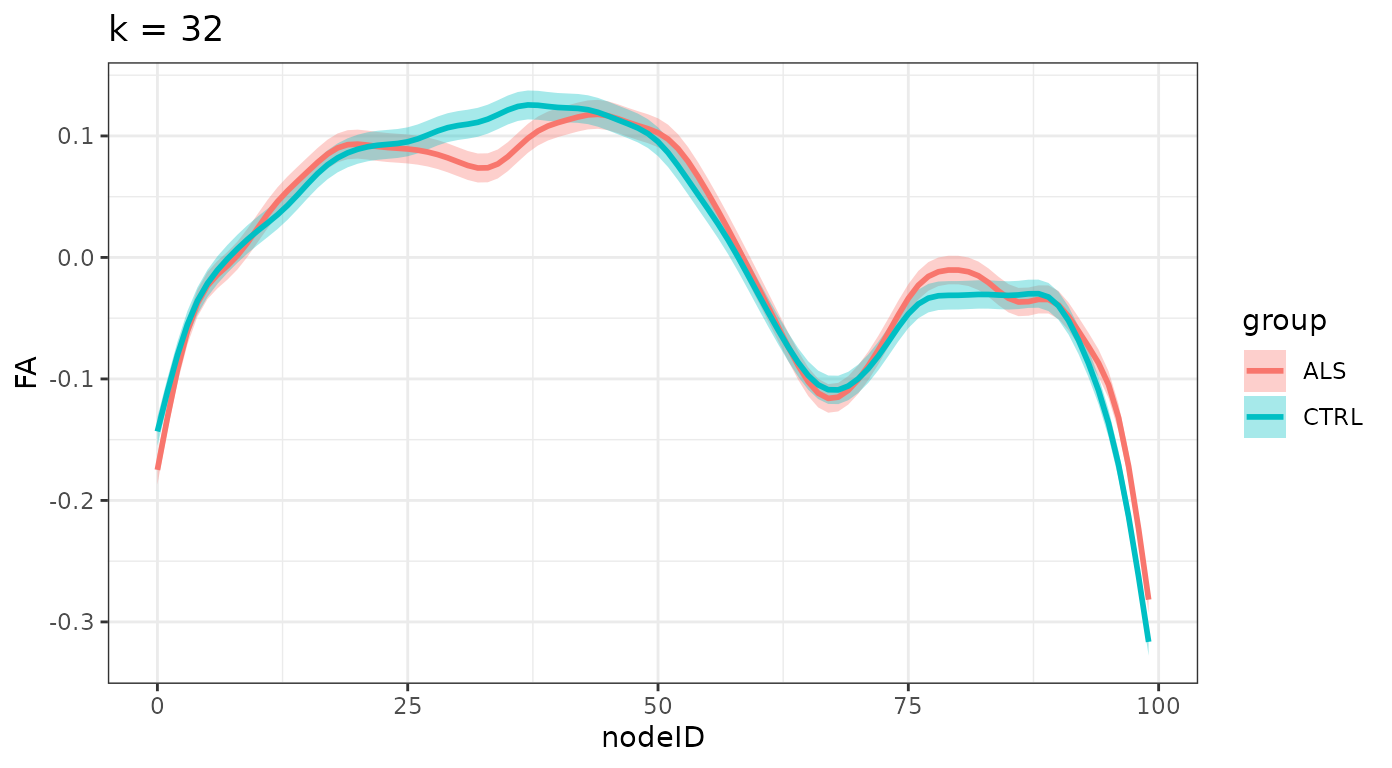
}And we plot the smooths, divided by group for each one of these levels:
plots <- list()
for (i in 1:length(k_values)){
plots[[i]] <- models[[i]] %>%
smooth_estimates() %>%
add_confint() %>%
dplyr::filter(.type != "Random effect") %>%
ggplot(aes(x = nodeID, y = .estimate, ymin = .lower_ci,
ymax = .upper_ci, group = group, color = group,
fill = group)) +
geom_ribbon(color = NA, alpha = 0.35) +
geom_line(linewidth = 1) +
scale_y_continuous(name = "FA") +
ggtitle(sprintf("k = %d", k_values[i])) +
theme_bw()
}
names(plots) <- sprintf("k = %d", k_values)
plots
#> $`k = 4`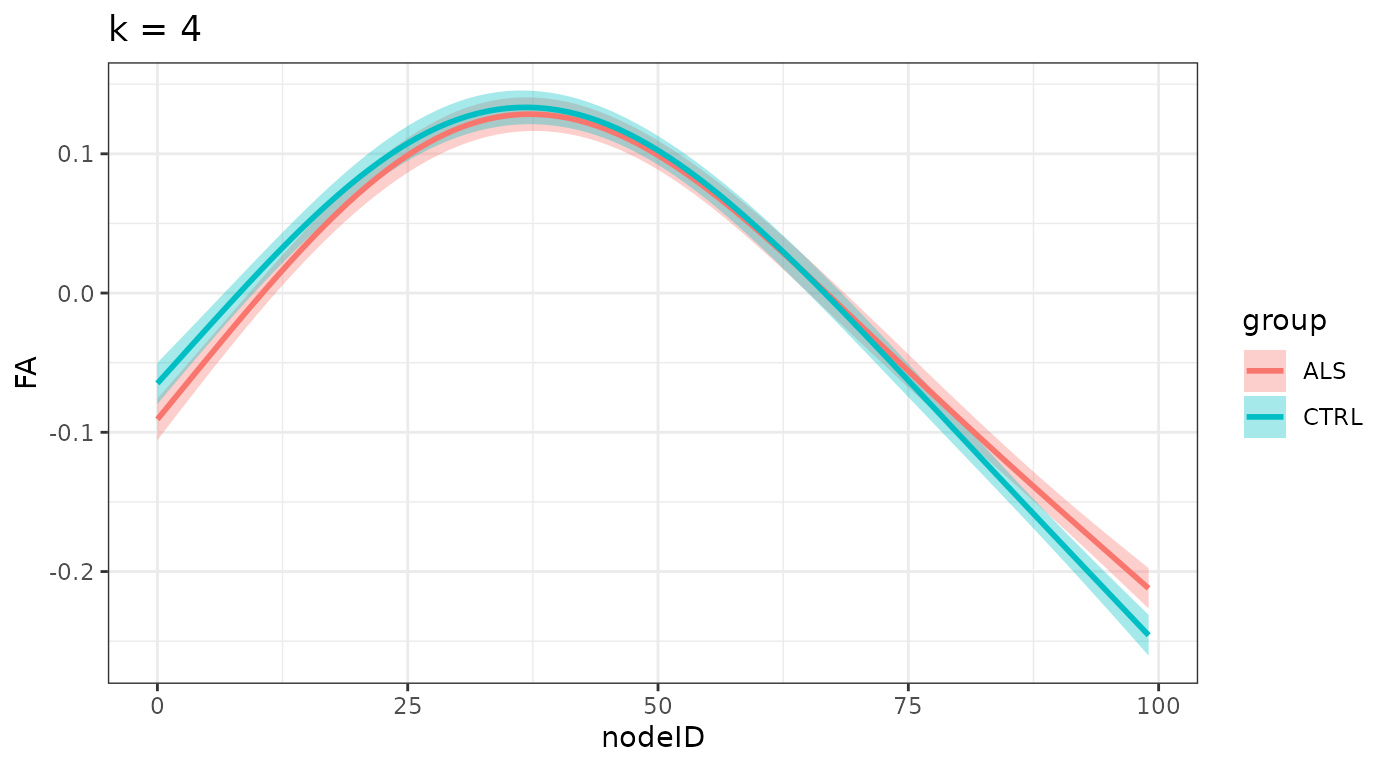
#>
#> $`k = 8`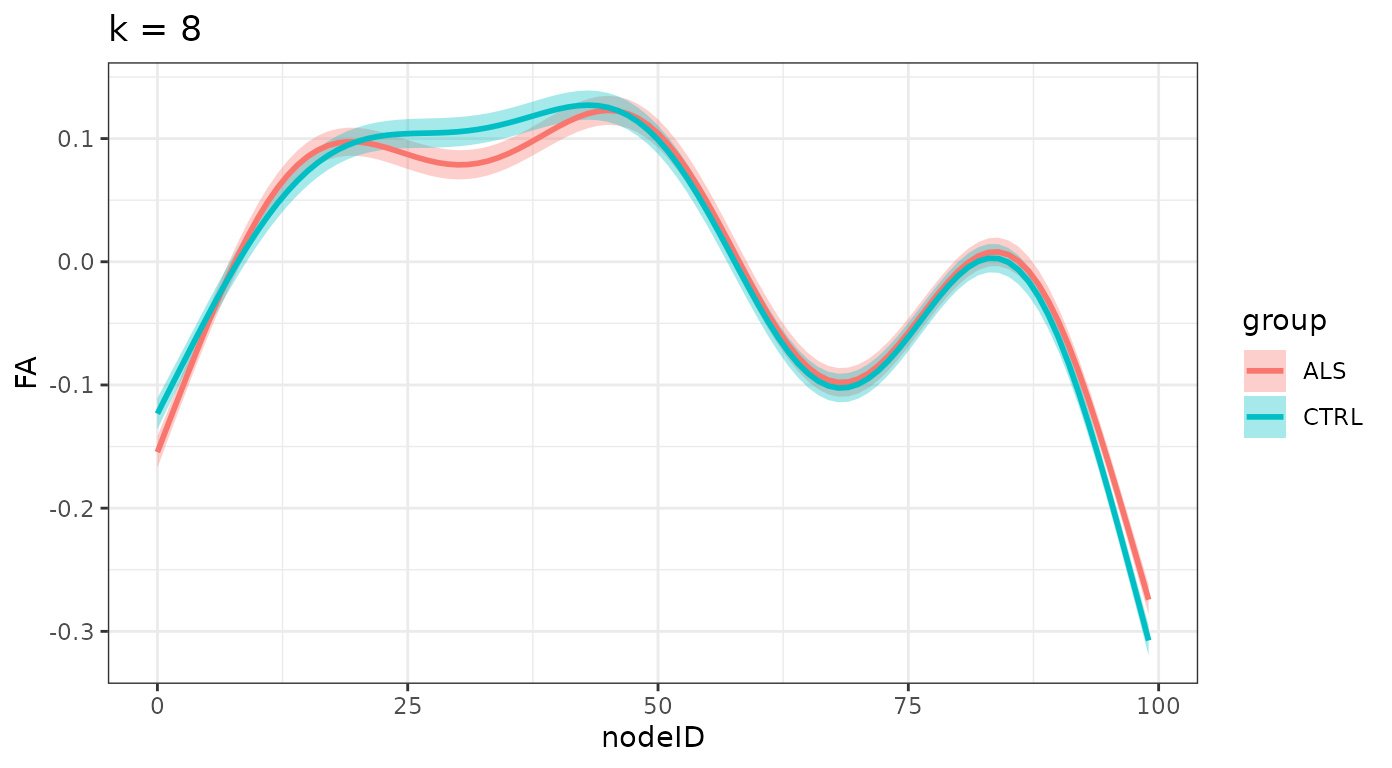
#>
#> $`k = 16`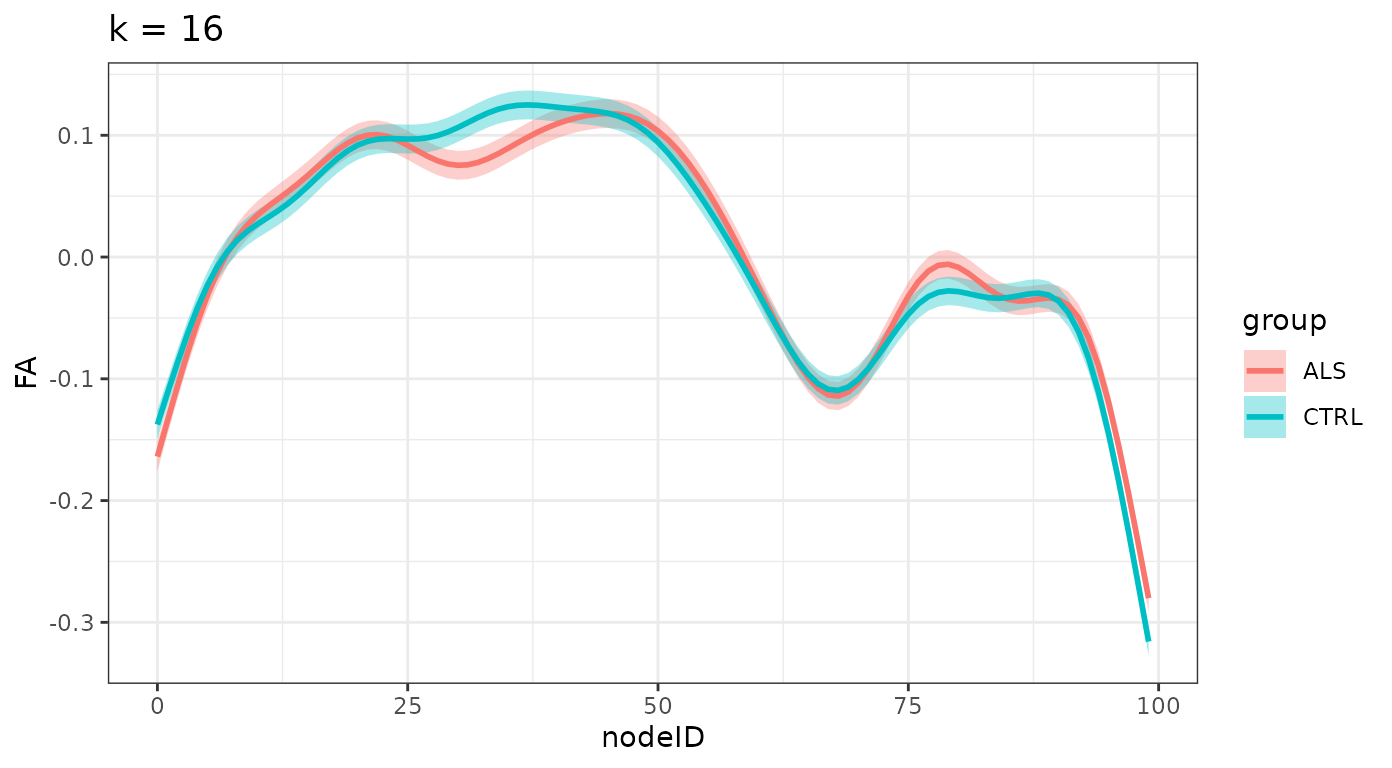
#>
#> $`k = 32`